If you have been reading through “Epigenetics vs Genetics” you will recall I said there is a great variety of molecules and mechanisms involved in epigenetic changes.
Here is a list of the most common mechanisms:
- DNA methylation
- Histone modifications
- Non-coding RNAs
- Genomic imprinting
- Silencing of the X chromosome
Now I will briefly explain the basics of the first two in the list.
DNA methylation
In chemistry, a methyl group is a small molecule that contains one carbon and three hydrogen atoms. All three hydrogen atoms are bound to the carbon. This is a very stable group and is found in many organic compounds.
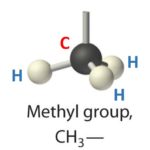
So DNA methylation is, as you surely have guessed, the addition of a methyl group to the DNA. But there are some rules. The methyl group cannot bind anywhere in the DNA, it is only added to the “letter” C (Cytosine).
The task of methylating DNA is performed by a group of enzymes called DNA methyltransferases (DNMTs).
Where do DNMTs get the methyl groups from? I mean, these enzymes are proteins put methyl groups in the DNA but they themselves do not have methyls, they need to “steal” them from other molecules. In fact, there are some generous molecules called “methyl donors” that donate their methyl groups to DNMTs, and they use an intermediary for doing so: a molecule called S-adenosylmethionine (SAM).
So, the transfer of methyl groups goes like this:

The methyl donor can be any compound with methyl groups, and there are many metabolites that serve as methyl donors. Interestingly, these metabolites are not only human metabolites but also microbial. In other words, our microbiota produce compounds that can lead to epigenetic changes in our DNA through methylation.
SAM has a methyl group in its composition. So, methyl donors, our human or microbial metabolites, are necessary in order to generate SAM.
Finally, SAM transfers its methyl group to a DNMT, the only enzyme capable of adding the methyl group to the cytosines (C) in DNA.
Histone modifications
Histones are proteins associated to DNA. They are very important to keep the DNA packaged so that it fits within the nucleus of the cell. Different types of histones form structures around of which DNA is wound. These structures are called nucleosomes. Think of them as balls with two loops of string of DNA around them.
Histones are also key players in epigenetic changes. They can be modified in many ways and these modifications affect how the genes near the histones are behaving: whether they are active, inactive or something in between.
The two most common modifications in histones are acetylation and methylation.
You have learned about DNA methylation above. Well, histone methylation is again the addition of methyl groups but this time to histones instead of DNA.
The specific epigenetic effect of methylating histones varies a great deal, depending on many factors like the particular histone that is methylated, the position within the histone, whether it gets one, two, or three methyl groups, the epigenetic changes that are nearby… It is complex and not for a basic introduction like this. Just keep the idea that histone methylation can mean either expression or repression of genes, depending on the factors mentioned above.
Moving on to acetylation…
If we have been talking about methyl groups in DNA and histone methylation, now we need to talk about acetyl groups. These are also found in many biological molecules and its chemical formula is CH3CO (notice that an acetyl group has a methyl group, CH3, within).
Simply, histone acetylation is the addition of acetyl groups to histone proteins. Acetylation is carried out by enzymes called histone acetyltransferases (HATs). Other enzymes do the opposite job, this is, remove acethyl groups from histones. These are called histone deacetylases (HDACs).
Again, acetylation and deacetylation of histones can cause a variety of epigenetic effects depending on where it is ocurring. But, as a general rule, acetylation leads to activation of genes and deacetylation to inactivation.